MNE Custom Filters
The internship I have right now just entails me to convert the toolbox they’ve created in MATLAB to Python. Obviously didn’t know this was going to happen but It definitely has its pros and cons. Con: I spend the entire day reading the documentations of SciPy, MNE, and MATLAB’s EEG Lab. Pro: Since I don’t know jack-shit converting someone else’s badly written code to another language is right up my alley.
The code I had used a Butterworth low-pass filter instead of MNE’s inbuilt.
A little bit of research and I guess the first thing you end up on is this, where oddly enough you’re greeted with a please shoo off this is for math-geniuses only but here’s the deal:
Filtering is essential, but it can lead to severe distortions if applied incorrectly.
~ An Introduction to ERP Technique
In this blog post, we’ll look into custom filters and how we can apply it using MNE.
The problem with using one of MNE’s inbuilt filters is that the roll-off is actually quite steep and this is quite visible
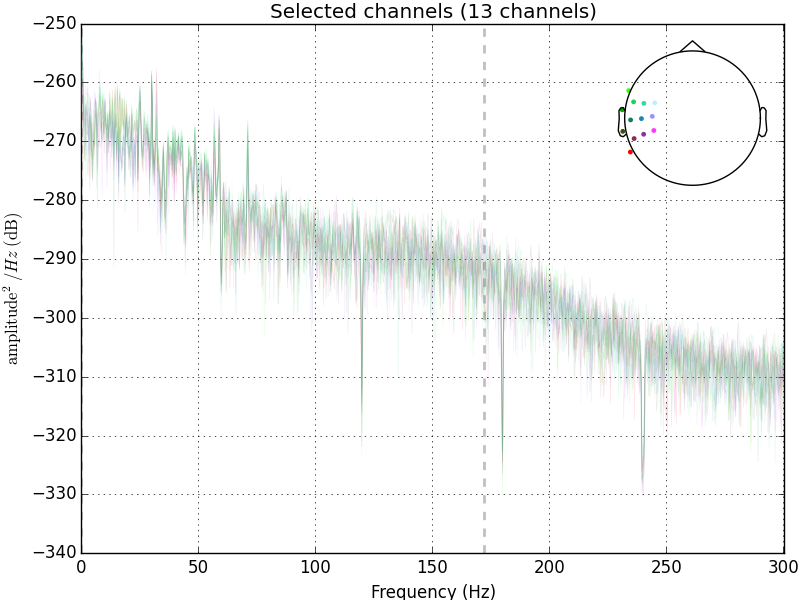
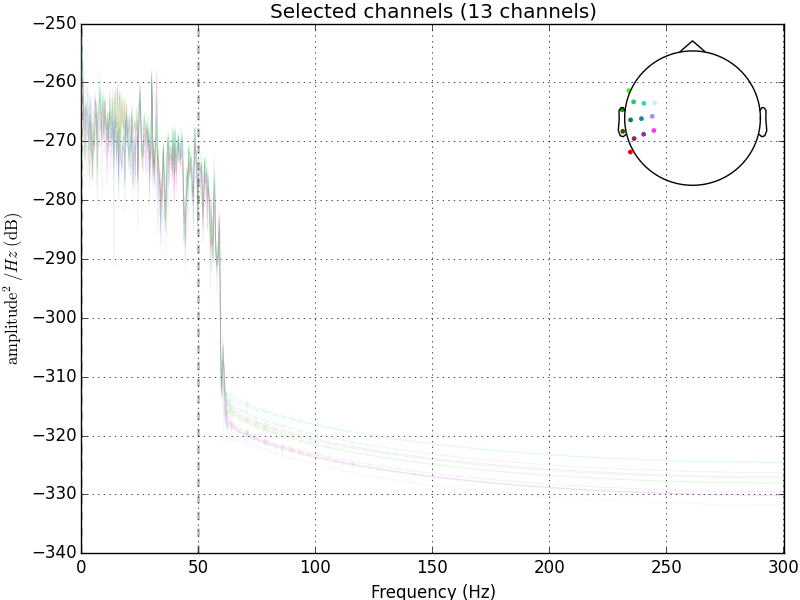
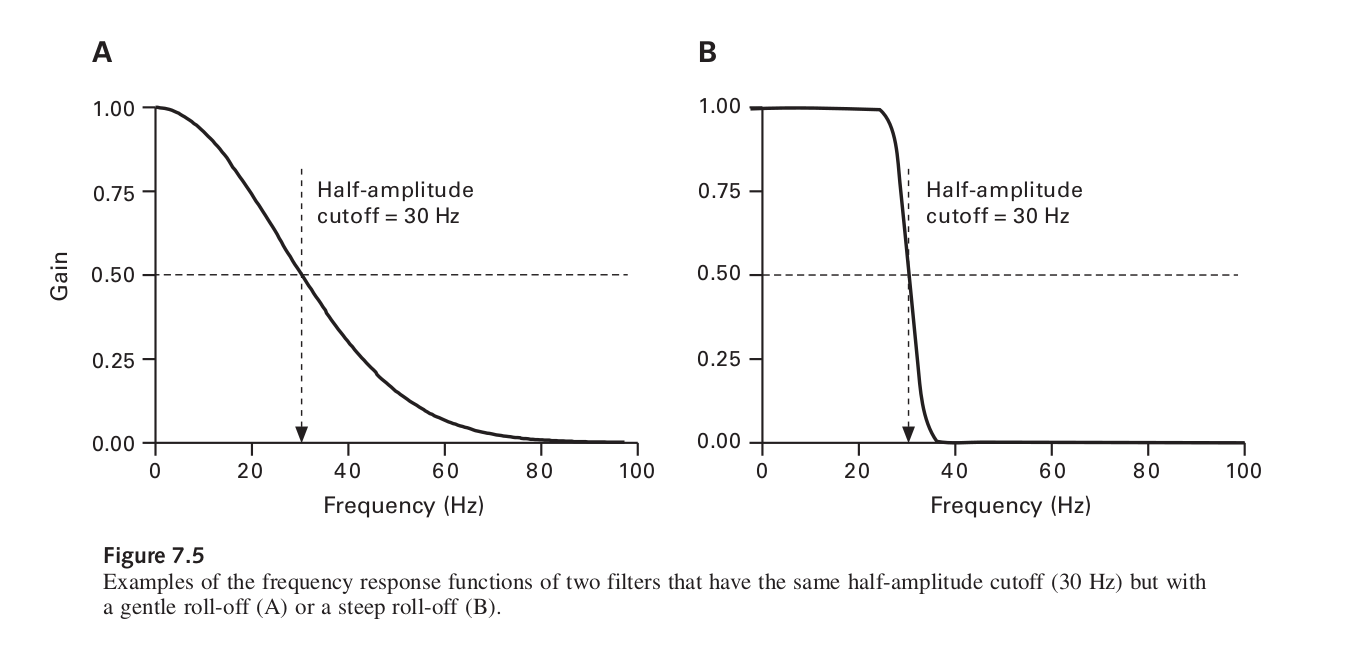
Edge artifacts can be created when the cutoff frequency is low (for both low- and high- pass filters) and when the roll-off is steep, because these factors require the use of more time points to compute the filtered data. Edge artifacts are not a problem when you have a long continuous EEG as the edge artifact will only be a very small part of it. Now it’s usually fine to apply low-pass filters to epoched EEG or averaged ERPs, but high-pass filters should usually be applied to the continuous EEG.
IIR or FIR?
IIR filters (e.g., Butterworth filters) do not need to use as many time points as FIR filters. Consequently, it can be advantageous to use an infinite impulse response filter when you are filtering epoched EEG data or averaged ERP data. For this reason, ERPLAB Toolbox mainly implements Butterworth filters.
With all that said, let’s implement our custom Butterworth and notch filters.
import matplotlib
import pathlib
import mne
import mne_bids
import os
import numpy as np
from numpy.fft import fft, fftfreq
from scipy import signal
from mne.time_frequency.tfr import morlet
from mne.viz import plot_filter, plot_ideal_filter
# matplotlib.use('TKAgg')
import matplotlib.pyplot as plt
import numpy as np
sample_data_folder = pathlib.Path('data/')
name_of_subject = input("Which subject do you want to analyse? with .edf appended")
sample_data_raw_file = os.path.join(sample_data_folder, name_of_subject)
print(sample_data_raw_file)
raw = mne.io.read_raw_edf(sample_data_raw_file)
print(raw.info, raw.info["ch_names"], raw.info["dig"])
<Info | 7 non-empty values
bads: []
ch_names: EEG Fp1-Ref, EEG Fp2-Ref, EEG F3-Ref, EEG F4-Ref, EEG C3-Ref, ...
chs: 43 EEG
custom_ref_applied: False
highpass: 0.0 Hz
lowpass: 500.0 Hz
meas_date: 2019-12-01 22:34:06 UTC
nchan: 43
projs: []
sfreq: 1000.0 Hz
> ['EEG Fp1-Ref', 'EEG Fp2-Ref', 'EEG F3-Ref', 'EEG F4-Ref', 'EEG C3-Ref', 'EEG C4-Ref', 'EEG P3-Ref', 'EEG P4-Ref', 'EEG O1-Ref', 'EEG O2-Ref', 'EEG F7-Ref', 'EEG F8-Ref', 'EEG T3-Ref', 'EEG T4-Ref', 'EEG T5-Ref', 'EEG T6-Ref', 'EEG Fz-Ref', 'EEG Cz-Ref', 'EEG Pz-Ref', 'POL E', 'POL PG1', 'POL PG2', 'EEG A1-Ref', 'EEG A2-Ref', 'POL T1', 'POL T2', 'POL X1', 'POL X2', 'POL X3', 'POL X4', 'POL X5', 'POL X6', 'POL X7', 'POL SpO2', 'POL EtCO2', 'POL DC03', 'POL DC04', 'POL DC05', 'POL DC06', 'POL Pulse', 'POL CO2Wave', 'POL $A1', 'POL $A2'] None
events_from_annot, event_dict = mne.events_from_annotations(raw)
Remove channels that we don’t have the mapping for
raw_removed = raw.copy().drop_channels(['POL E', 'POL PG1', 'POL PG2', 'EEG A1-Ref', 'EEG A2-Ref', 'POL T1', 'POL T2', 'POL X1', 'POL X2', 'POL X3', 'POL X4', 'POL X5', 'POL X6', 'POL X7', 'POL SpO2', 'POL EtCO2', 'POL DC03', 'POL DC04', 'POL DC05', 'POL DC06', 'POL Pulse', 'POL CO2Wave'])
Re-name to Re-map
The renaming
- t3 -> T7
- t4 -> T8
- t5 -> P7
- t6 -> P8
mapping = {'EEG Fp1-Ref': 'Fp1', 'EEG Fp2-Ref': 'Fp2', 'EEG F3-Ref':'F3',
'EEG F4-Ref':'F4', 'EEG C3-Ref':'C3', 'EEG C4-Ref':'C4', 'EEG P3-Ref':'P3',
'EEG P4-Ref':'P4', 'EEG O1-Ref':'O1', 'EEG O2-Ref':'O2', 'EEG F7-Ref':'F7',
'EEG F8-Ref':'F8', 'EEG T3-Ref':'T7', 'EEG T4-Ref':'P8', 'EEG T5-Ref':'P7',
'EEG T6-Ref':'T8', 'EEG Fz-Ref':'Fz', 'EEG Cz-Ref':'Cz', 'EEG Pz-Ref':'Pz',
'POL $A1':'A1', 'POL $A2':'A2'}
raw_rename = raw_removed.rename_channels(mapping)
raw_no_ref_a1a2 = raw_rename.copy().drop_channels(['A1', 'A2'])
raw_no_ref_a1a2.plot(duration=0.5)
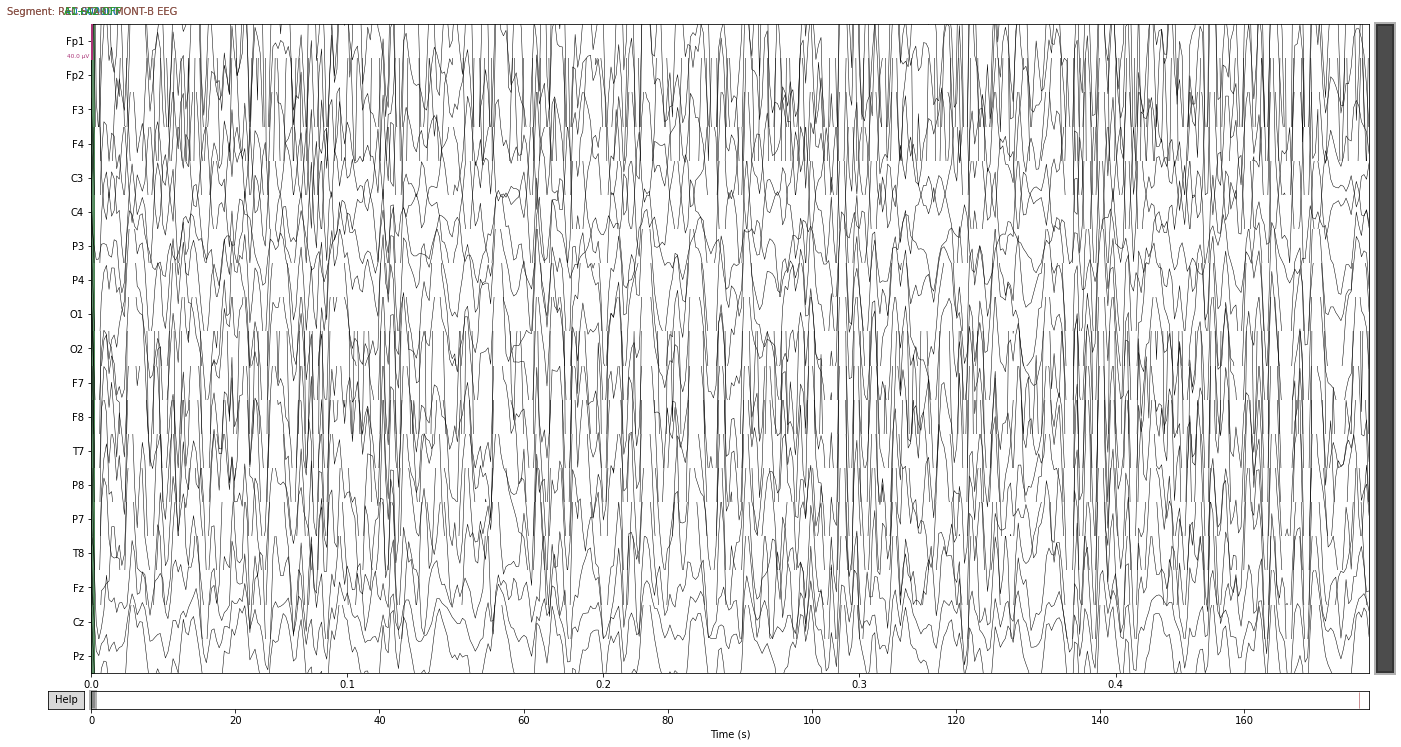
montage = mne.channels.read_custom_montage('Standard-10-20-Cap19.locs')
raw_montage = raw_no_ref_a1a2.copy().set_montage(montage, match_case=False)
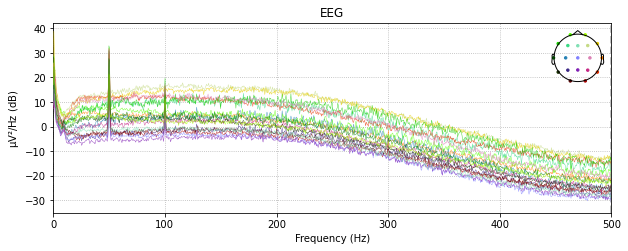
raw_montage.plot_psd()
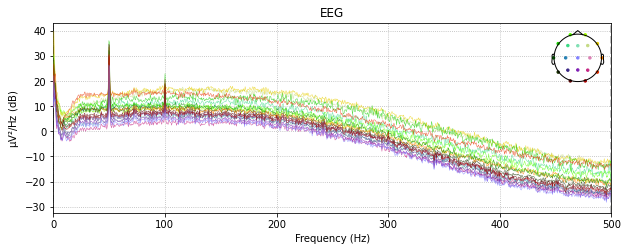
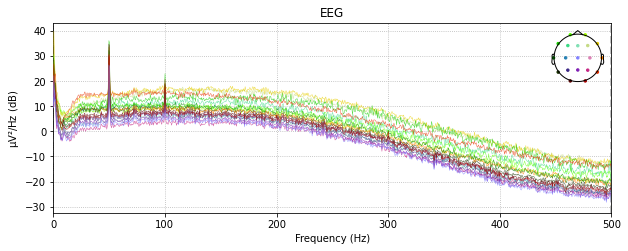
Removing and reconstruct by spline interpolation
Bad channels if not redeemable else flagged portions are removed
raw_loaded = raw_montage.copy().load_data()
raw_loaded.interpolate_bads(reset_bads=False, verbose=False)
raw_loaded.plot_psd()
raw_loaded.plot(duration=0.5)
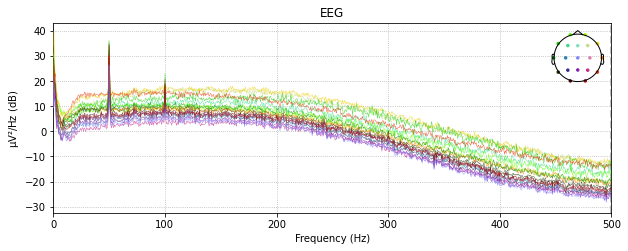
Re-reference the Signal: Average Reference
Averaging is a linear operation. Consequently, it doesn’t matter if you filter the EEG immediately before averaging or if you filter the ERP immediately after averaging. Re-referencing is also a linear operation (except in unusual cases), so you can filter either before or after you re-reference the data.
raw_avg_ref = raw_loaded.load_data().copy().set_eeg_reference(ref_channels='average')
raw_avg_ref.plot(duration=0.5)
raw_avg_ref.plot_psd()
EEG channel type selected for re-referencing
Applying average reference.
Applying a custom EEG reference.
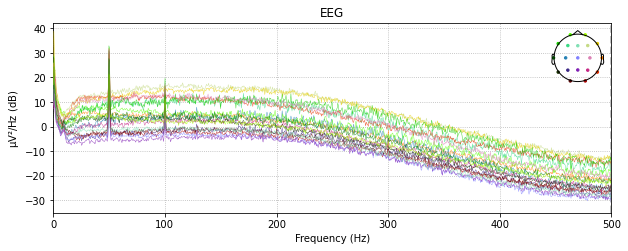
Notch Filters
picks = raw_loaded.info["ch_names"]
def notch_bandpass(line_noise, sfreq, notch_width, index):
w = line_noise/(sfreq/2); # 50 Hz line noise in radians
# print(notch_width)
notch_width = notch_width/(sfreq/2);
b, a = signal.iirnotch(w*index, notch_width)
return b,a
def notch_filter(sig, line_noise, sfreq, notch_width, index):
m = sig[0]
# n = sig[1]
# print("m:",m)
# o = np.array((m,n))
# print("o:",o)
b,a = notch_bandpass(line_noise, sfreq, notch_width, index)
c = signal.filtfilt(b, a, m, padlen=0)
# print("c:",c)
return c
Butterworth High
def butter_bandpass_high(butterworth_order, h_freq, sfreq):
b, a = signal.butter(butterworth_order, h_freq/(sfreq/2),'high')
return b,a
def butter_filter_high(sig, butterworth_order, h_freq, sfreq):
m = sig[0]
b,a = butter_bandpass_high(butterworth_order, h_freq, sfreq)
c = signal.filtfilt(b, a, m, padlen=0)
return c
Butterworth Low
def butter_bandpass_low(butterworth_order, l_freq, sfreq):
b, a = signal.butter(butterworth_order, l_freq/(sfreq/2),'low')
return b,a
def butter_filter_low(sig, butterworth_order, l_freq, sfreq):
m = sig[0]
b,a = butter_bandpass_low(butterworth_order, l_freq, sfreq)
c = signal.filtfilt(b, a, m, padlen=0)
return c
Now applying all filters
def prepsig(raw_edf=raw_avg_ref, signals=picks, notch_width=5, butterworth_order=5, sfreq=1000, h_freq=1.0, l_freq=125, line_noise=50, initial_transient=0, final_transient=4):
# Sig = prepsig(Sig, bw, ord, fs, fhi, Ln, Ti, Tf)
# function with full input argument
# Defaults
# notch_width ... Bandwith of notch Filter: default 5 Hz
# butterworth_order ... butterworth Filter Order: default 5
# s_freq ... Sampling frequency: default 250 Hz
# h_freq ... Cut-off frequency for removing slow drift: default 0.53 Hz
# l_freq ...Lowpass
# line_noise ... Line-noise specification: default 50 Hz
# initial_transient ... Initial transient: default 4 s
# final_transient ... Final transient: default 0 s
# goes through 9 times for all nineteen signals
# pick a signal
raw_edf = raw_edf.copy().load_data()
for index in range(1, 10):
for sig in range(0, 19):
# a = butter_bandpass_filter(raw_edf[x], line_noise, sfreq, butterworth_order, index)
# print(type(a))
raw_edf[sig] = notch_filter(raw_edf[sig], line_noise, sfreq, notch_width, index)
# raw_edf[sig] = butter_bandpass_filter_1(raw_edf[sig][0], line_noise, sfreq, butterworth_order, index)
print("Notch filter applied")
raw_edf.plot_psd()
for sig in range(0, 19):
raw_edf[sig] = butter_filter_high(raw_edf[sig], butterworth_order, h_freq, sfreq)
print("Butterworth High")
raw_edf.plot_psd()
for sig in range(0, 19):
raw_edf[sig] = butter_filter_low(raw_edf[sig], butterworth_order, l_freq, sfreq)
print("Butterworth Low")
raw_edf.plot_psd()
return raw_edf
raw_filt = prepsig()
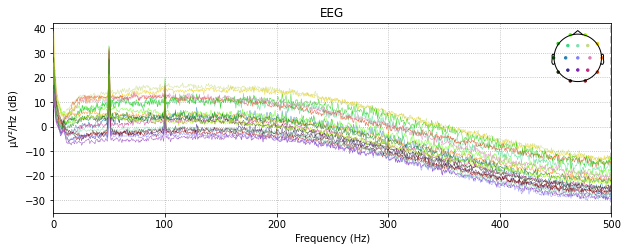
Butterworth High
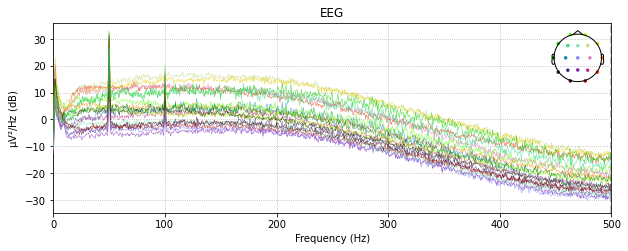
Butterworth Low
# what we want
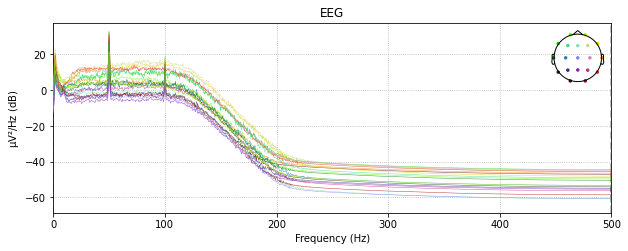
# low pass filtering below 50 Hz # raw_filt or raw_notch
raw_low_pass = raw_avg_ref.copy().filter(None, 50., fir_design='firwin')raw_low_pass.plot_psd()
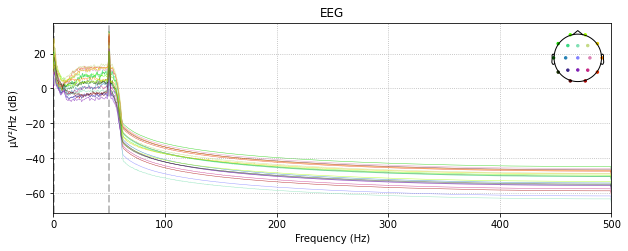
See the difference?
